What is this wiki about?¶
This wiki presents the code used by the CP3-llbb group, to perform analyses of p-p collisions collected by the CMS experiment
What are the prerequisites?¶
If you intend to use the code in its vanilla version, you need:
- an access to ingrid, see the CP3-IT FAQ to get some more info
- some knowledge of CMSSW, the CMS software
If you intend to develop and follow what other do, it is a (very recommended) good idea to:
- follow the llbb group meetings
- register to the cp3-llbb CERN egroup
- have some knowledge about using git:
- this online book is a nice one
- this five-minutes introduction presents the workflow we use for developing code
- ask questions!
- do not be shy of posting issues if you notice what you think is a bug
- do not hesitate to make suggestions about improving any documentation if things are not clear

There is plenty of repositories here, what do I do?¶
Specific instructions on how to install and use them should be in the README of each repository (if not, do complain). The flow is the following:
-
CMSSW code (The technical documentation is not yet ready (September 2015)):
- the Framework is the core of the code, it transforms MINIAOD objects into non-flat ROOT trees containing the reconstruced leptons, MET, (b)jet, and more. It applies the high-level corrections needed by everyone (leptons SF, JES, JEC, etc.) and also constructs some more high-level objects (dileptons). It can be run stand-alone, but is intended to be used with an analysis (see below). The code is in the Framework repository, and there is also Doxygen reference documentation
- the analyses (HHAnalysis, TTAnalysis, ZAAnalysis) are CMSSW plugins. The intent of these is to run the framework and gather more info in the trees specific to each analysis (MC truth, reconstruction of the top, the Higgs boson, the Z boson). Except if you want to produce very basic control plots, this is probably what you want to run.
-
using the grid:
- now that we have some CMSSW code compiling and running, you probably want to run it on the grid, with CRAB3, in order to create the trees for many MC and data samples, to perform some analysis
- GridIn is our CMSSW package to easily do so: it prepares the crab config files so that you just have to launch them
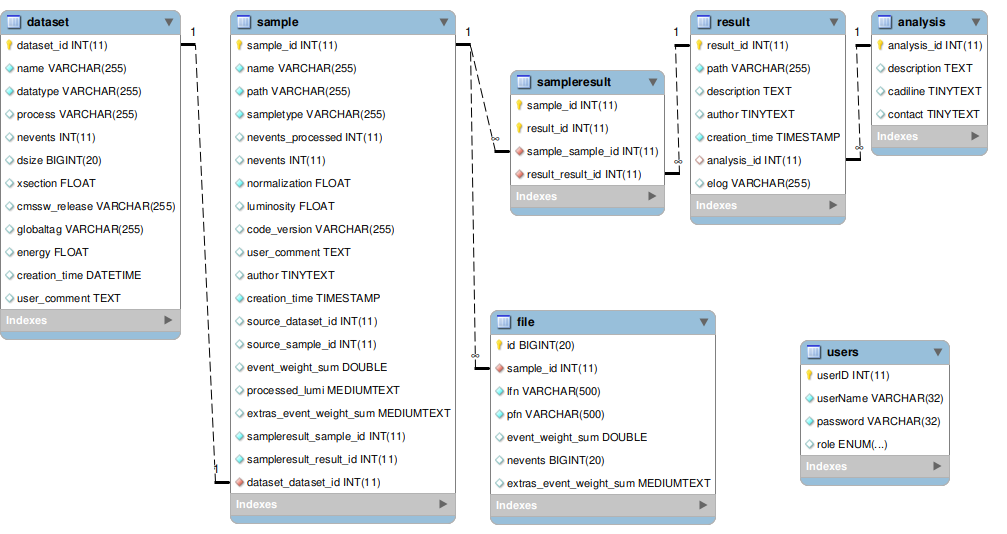
- if your grid jobs are successful, you want to write down somewhere what is it you've down. To do so we use SAMADhi, our local database (there is also a web interface), to bookkeep what code has been run to produce the trees and where they are stored. The format and links of the database are the displayed below.
-
produce some plots:
- now that you have trees you can make plots. You can use your own code to do so, but given some trees are quite big, we have some utilities to help
- CommonTools contain helpers, in particular histFactory is here to mass-produce histogram files
- now that you have histogram files for all your samples, plotIt is a nice utility to stack them and display them in the latest recommended CMS style